-
Latest Version
-
Operating System
Windows 7 / Windows 7 64 / Windows 8 / Windows 8 64 / Windows 10 / Windows 10 64
-
User Rating
Click to vote -
Author / Product
-
Filename
snapgene_5.1.5_win.exe
-
MD5 Checksum
ab038cb415f24bbe7dfaf70a62fb6749
Sometimes latest versions of the software can cause issues when installed on older devices or devices running an older version of the operating system.
Software makers usually fix these issues but it can take them some time. What you can do in the meantime is to download and install an older version of SnapGene 5.1.5.
For those interested in downloading the most recent release of SnapGene or reading our review, simply click here.
All old versions distributed on our website are completely virus-free and available for download at no cost.
We would love to hear from you
If you have any questions or ideas that you want to share with us - head over to our Contact page and let us know. We value your feedback!
What's new in this version:
New Functionality:
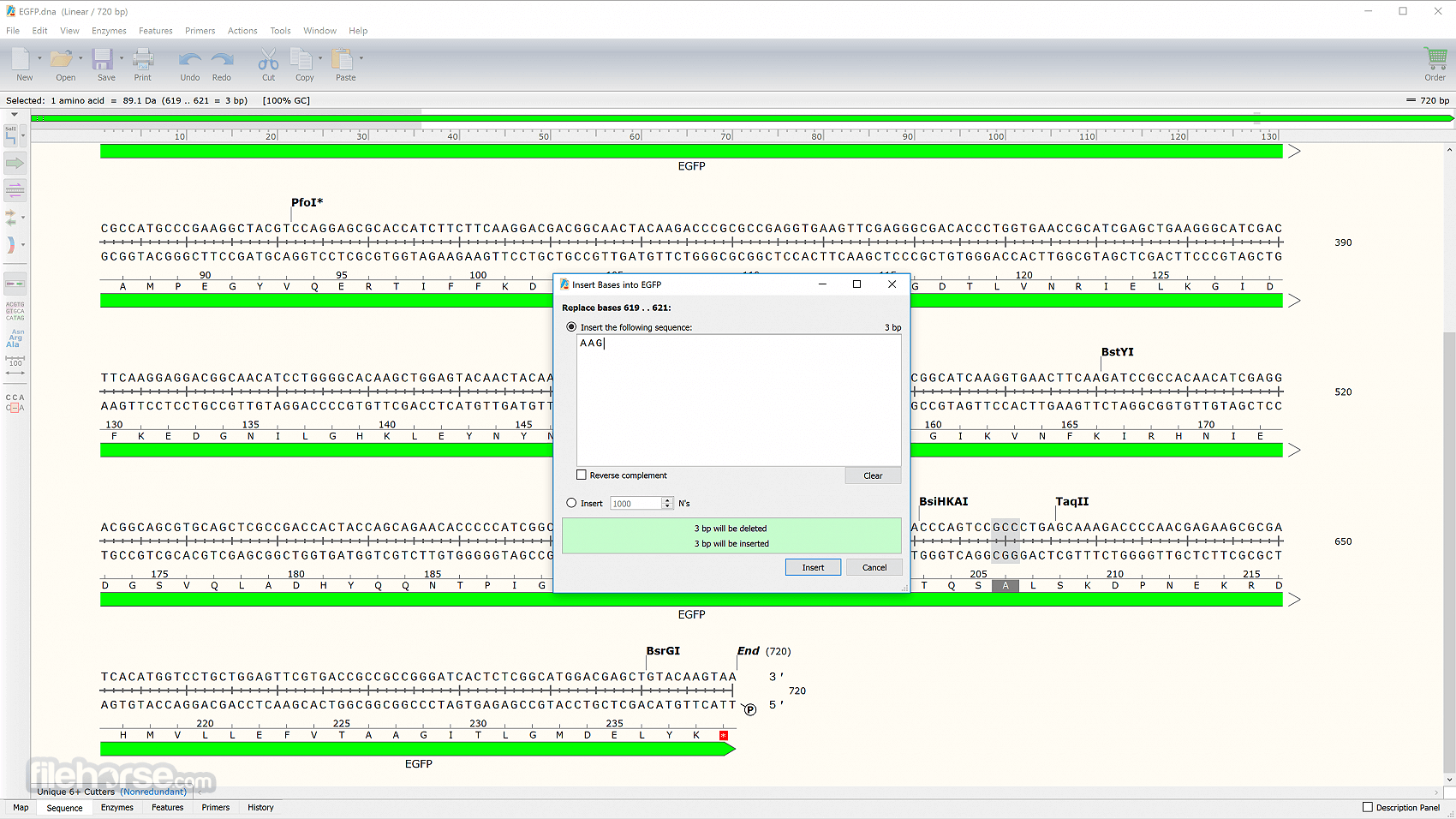
- Added a "Copy Rich Text" command for selections in alignments, to provide the option of copying either simple sequences or sequences with metadata
Enhancements:
- Added Invitrogen's "pScreen-iT LacZ-Dest" to the list of Gateway® Destination vectors
- Modified the statistics in pairwise alignments to show two digits after the decimal point instead of one
- Added time estimates to various progress dialogs
- Improved the order of "Copy" actions in the Edit menu
- Made various textual, alignment, and spacing improvements
- Enabled export of a map or history while viewing any tab
Fixes:
- Ensured reliable import of primers copied to the clipboard using Microsoft Office
- Added support for dragging and dropping FASTA archives into the Assemble Contigs dialog
- Preserved zoom and split view display options when switching between files in a collection
- Enhanced the Gene Construction Kit importer to capture the full set of notes in the "General Info" section
- Improved stability when computing and viewing multiple sequence alignments
- Corrected an issue that could cause features to be erroneously detected around the numerical origin of a linear sequence
- Ensured that proper file extensions were included when batch converting files from one format to another
- Ensured correct setting of the default button in the Find controls when pressing and releasing Shift in a sequence trace window
- Corrected a regression with the navigation buttons when viewing an alignment to a reference sequence
- Addressed issues with the purple bar and the Tm column when importing primers from another file
- Corrected the displayed molecular weight when adding a translated feature to the common features database
- Removed the colors button in cloning dialogs, and streamlined the side toolbar in the Edit DNA Ends, Browse Common Features, and Mutageneis dialogs
- Improved the display of long sequence names within circular maps
- Corrected a regression by removing cut locations for ancestral restriction sites in History view
- Removed the inappropriate "Preserve feature annotations" control from the New File dialog, and the inappropriate "Detect common features" control when inserting or replacing bases in a sequence trace window
- Improved stability when assembling contigs using FASTQ data
- Disabled the Show/Hide All Enzymes commands when viewing protein files
- Corrected an issue in which the endpoints of a selection were not updated in the selection bar after renumbering the origin of a linear sequence
- Fixed an issue that prevented immediately using SnapGene without restarting after activating a Flexera-based shared license
- Corrected an issue with computing % GC when partially degenerate residues (B, D, H, and V) were present
- Ensured that only the zoomed region is shown for the root map in History view
- Addressed an issue in which the Save As dialog would vanish immediately when attempting to choose a different name instead of saving over an existing file
- Ensured that enzyme set menus are refreshed after using Manage Enzyme Sets
- Ensured that the desired endpoint modifications are correctly applied when designing a synthetic construct
- Improved the registration of file associations on macOS
 OperaOpera 118.0 Build 5461.41 (64-bit)
OperaOpera 118.0 Build 5461.41 (64-bit) PC RepairPC Repair Tool 2025
PC RepairPC Repair Tool 2025 PhotoshopAdobe Photoshop CC 2025 26.5.0 (64-bit)
PhotoshopAdobe Photoshop CC 2025 26.5.0 (64-bit) OKXOKX - Buy Bitcoin or Ethereum
OKXOKX - Buy Bitcoin or Ethereum iTop VPNiTop VPN 6.4.0 - Fast, Safe & Secure
iTop VPNiTop VPN 6.4.0 - Fast, Safe & Secure Premiere ProAdobe Premiere Pro CC 2025 25.2.1
Premiere ProAdobe Premiere Pro CC 2025 25.2.1 BlueStacksBlueStacks 10.42.51.1001
BlueStacksBlueStacks 10.42.51.1001 Hero WarsHero Wars - Online Action Game
Hero WarsHero Wars - Online Action Game SemrushSemrush - Keyword Research Tool
SemrushSemrush - Keyword Research Tool LockWiperiMyFone LockWiper (Android) 5.7.2
LockWiperiMyFone LockWiper (Android) 5.7.2









Comments and User Reviews