-
Latest Version
SnapGene 8.0.3 LATEST
-
Review by
-
Operating System
Windows 7 / Windows 7 64 / Windows 8 / Windows 8 64 / Windows 10 / Windows 10 64
-
User Rating
Click to vote -
Author / Product
-
Filename
snapgene_8.0.3_win.exe
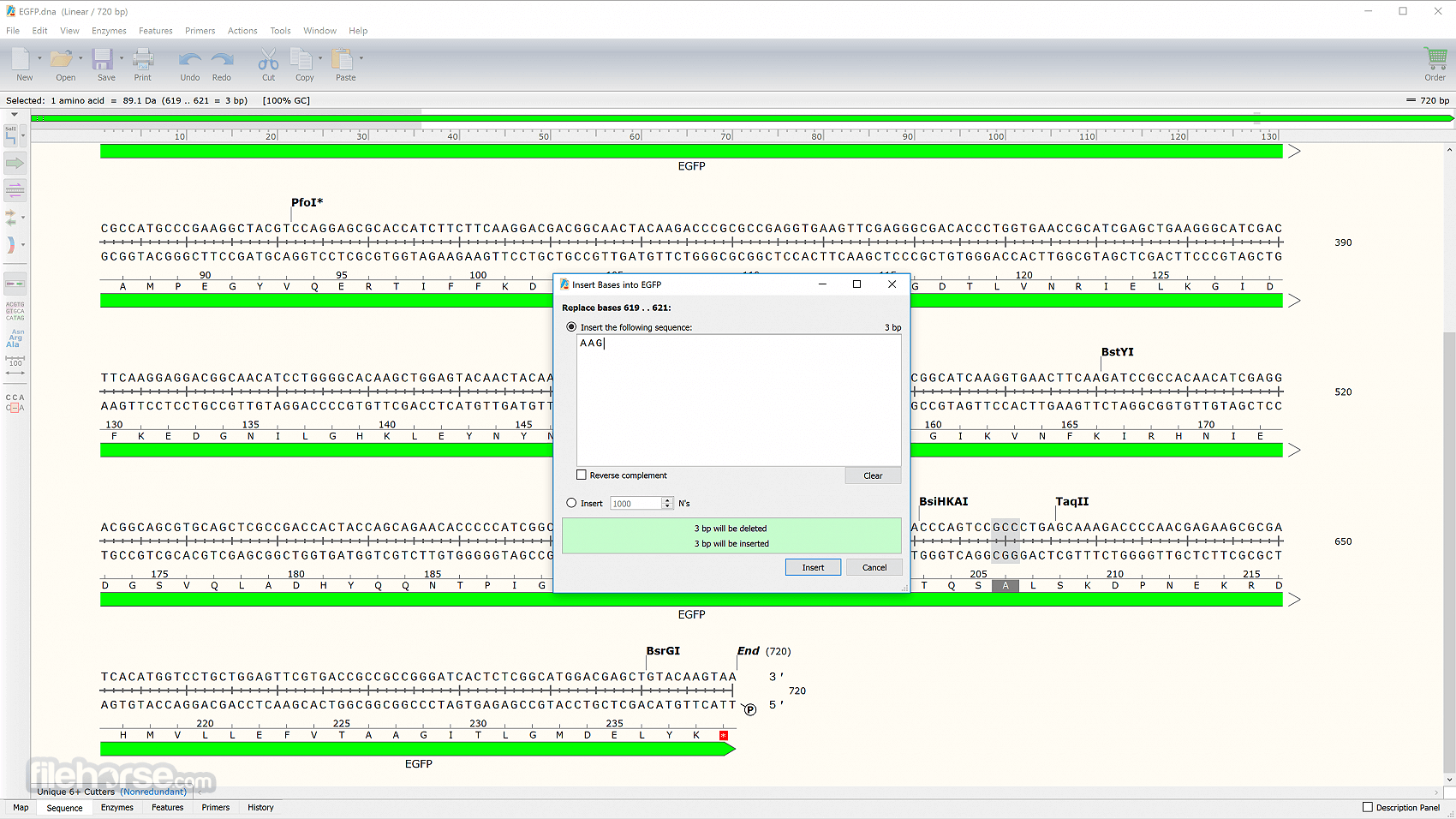
Take advantage of SnapGene’s efficient data handling to scan large DNA sequences with thousands of annotated features. Make insertions, deletions, replacements, and case changes. When a sequence is copied and pasted, features are automatically transferred. Annotate common features automatically, or annotate novel features manually.
Find common features in your DNA sequence using SnapGene’s extensive database. Additional features of your choosing can be added to a custom database.
It provides elegant, information-rich windows for simulating a variety of common cloning and PCR methods. Highlight unique restriction sites in bold font, or choose the automatically defined Unique Cutters or Unique 6+ Cutters enzyme set.
Use your own primers, or ask the app to design primers automatically. The product file stores the template and primers in its history. Assemble up to eight fragments. Select the fragments to be joined and their orientations, and Snap Gene will design primers.
Use a Sequence view to see at a glance whether two translated features are in the frame. If so, the translations are linked on the same line. If not, the translations are on separate lines. Use the powerful alignment tool to check whether an actual construct matches the simulated construct.
It automatically records operations to create a graphical history, and stores the ancestor constructs in the final file. Use the familiar, secure operating system of your computer to store and organize your Snap Gene files.
Export a sequence to GenBank or FASTA format. Export a map or simulated agarose gel to common image formats. Convert a sequence, map, or gel image to standard formats for use with other software.

The open exchange of information is crucial, so SnapGene and SnapGene Viewer provide options for reading and exporting common file formats.
How to Use
- Download SnapGene from the official website
- Install the software on your Windows PC
- Launch the application after installation
- Create or open a DNA sequence file
- Annotate features and edit sequences
- Simulate cloning procedures
- View plasmid maps and sequence details
- Export data or save your project
- Share files with collaborators
- Use tutorials for advanced features

Pricing
Academic Subscription: For universities and nonprofits using SnapGene for academic research. Annual pricing depends on the number of users — $350 for 1 user, $650 for 2, $1,625 for 5, $3,000 for 10. Includes all platform licenses, updates, and license management tools.
Corporate Subscription: For commercial use, priced higher — $1,845 for 1 user, $3,690 for 2, $9,225 for 5, $18,450 for 10 users per year. Offers the same benefits as the academic plan but for business environments.
Student Subscription: Available to enrolled students (undergrad, grad, postdoc, med residents) for $149/year. Must be purchased online by the individual student. Intended for academic work only.
Permanent Licenses: One-time purchase option with no updates or support. $2,500 for academic users and $9,600 for corporate. Not recommended due to lack of upgrades or compatibility guarantees.
Course Licenses: Free temporary licenses for educators offering SnapGene to students as part of a class.
All paid plans support installation on Windows, Mac, and Linux, and offer user management, deployment tools, and activation control. Corporate and Academic plans also support prorated additions and automatic renewal.

System Requirements
Operating System: Windows 11 or Windows 10 (64-bit)
Processor: 2.4 GHz or faster
RAM: Minimum 4 GB (8 GB recommended)
Hard Disk Space: At least 500 MB available
Display: 1280x800 resolution or higher
Internet: Required for activation and updates
PROS
- User-friendly interface
- Accurate cloning simulation
- Real-time plasmid visualization
- Broad file format support
- Extensive annotation tools
- Expensive licensing cost
- Limited free version features
- No native Linux support
- High resource consumption
- Steep learning curve for some users
What's new in this version:
Fixed:
- Restore missing option to print the Info panel
- Fixed an issue that prevented showing the full list of features and primers after toggling off zoom mode
- Fixed an issue that could prevent finding some primer binding sites or matches for searches that wrap around the numerical origin
- Fixed an issue where double clicking a File or otherwise attempting to start SnapGene a second time would eventually require activating again
- Fixed opening some .apr VectorNTI alignment files
- Updated various libraries (libemf, pcre2, zlib, Minizip, OpenSSL, xz) to the latest versions
 OperaOpera 118.0 Build 5461.41 (64-bit)
OperaOpera 118.0 Build 5461.41 (64-bit) PC RepairPC Repair Tool 2025
PC RepairPC Repair Tool 2025 PhotoshopAdobe Photoshop CC 2025 26.5.0 (64-bit)
PhotoshopAdobe Photoshop CC 2025 26.5.0 (64-bit) OKXOKX - Buy Bitcoin or Ethereum
OKXOKX - Buy Bitcoin or Ethereum iTop VPNiTop VPN 6.4.0 - Fast, Safe & Secure
iTop VPNiTop VPN 6.4.0 - Fast, Safe & Secure Premiere ProAdobe Premiere Pro CC 2025 25.2.1
Premiere ProAdobe Premiere Pro CC 2025 25.2.1 BlueStacksBlueStacks 10.42.51.1001
BlueStacksBlueStacks 10.42.51.1001 Hero WarsHero Wars - Online Action Game
Hero WarsHero Wars - Online Action Game SemrushSemrush - Keyword Research Tool
SemrushSemrush - Keyword Research Tool LockWiperiMyFone LockWiper (Android) 5.7.2
LockWiperiMyFone LockWiper (Android) 5.7.2






Comments and User Reviews